Ashwin Srinath and Linh Bao Ngo, Clemson University, August 29, 2018
Researchers from the Getman Group at Clemson University needed to post-process results from large-scale molecular dynamics simulations. The custom sequential Python scripts they had been using for this purpose were taking well over 72 hours (the limit for jobs on Clemson’s Palmetto HPC Cluster). Clemson University ACI-REFs at Clemson University Dr. Linh Ngo and Ashwin Srinath helped the researchers develop a post-processing pipeline that exploits Clemson’s Hadoop/Spark cluster (Cypress), bringing down the time for post-processing from a few days to under an hour.
The Objective
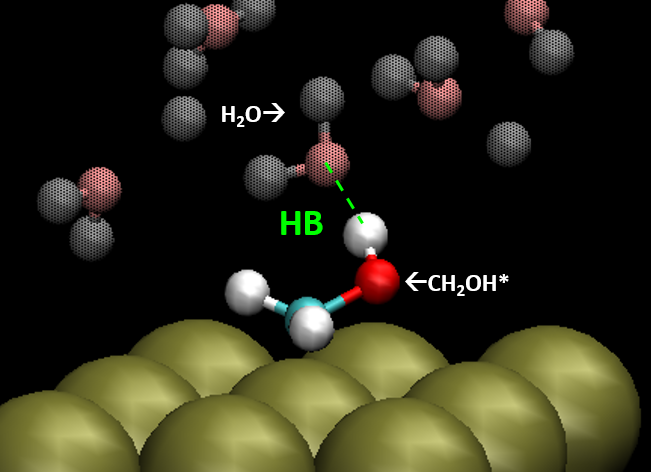
The Getman Group uses particle-based models to study the mechanisms of chemical reactions involving catalysts. They run large-scale molecular dynamics simulations on HPC using software such as LAMMPS (Large-scale Atomic/Molecular Massively Parallel Simulator) to generate particle trajectories. The resulting trajectory files contains hundreds of thousands of “frames,” each frame representing particle coordinates at a particular instant of time. Figure 1 shows an example of a typical frame of the trajectory file. For this particular analysis, they were interested in the number of Hydrogen bonds (HB) in each frame, and the averaged HB (AvgHB) for the entire simulation.
Previously, the group developed a Python program that read the trajectory file sequentially, i.e., “frame-by-frame,” counting the number of HB in for each frame and thenfinally computing the average. Since this was taking prohibitively long (longer than allowed on Palmetto Cluster) for larger simulation results, they approached Clemson ACI-REFs for help with speeding up their Python program. Our initial objective was to identify and eliminate any bottlenecks in the code, but after working with the group more, we suggested exploring other approaches, including using third-party libraries for performing post-processing and leveraging technologies such as Hadoop and Spark for handling the large amounts of data.
The Solution
1. Improving the original code
The initial effort involved studying the original Python program that the group was using, to determineand factors that influenced its performance. For this, we helped the group perform small benchmarks on smaller datasets (using fewer frames). It was quickly apparent that I/O (reading and/or writing data) from the disk was the limiting factor., The code spent most of its time reading data from the trajectory file and very little time doing actual computation. From walking through the code with the researchers, we discovered that at each time step (i.e., for each frame), the code was reading the entire trajectory file, rather than only the information relevant to that time step. Our immediate recommendation was to read the trajectory file block-by-block (about 3000 frames at a time) into memory to minimize the amount of I/O. This improved performance considerably, but the code still performed slowly, taking about 12 hours to analyze a 10 GB trajectory file.
2. Leveraging third-party packages for post-processing
Our next course of action was to explore the possibility of using existing, community-developed tools for performing the trajectory analysis, and possibly benefiting from the optimization efforts of other researchers doing similar work. We found the MDTraj library (https://github.com/mdtraj/mdtraj) to be the best candidate, as it (1) is in Python, (2) contains several Hbond analysis functions (http://mdtraj.org/1.6.2/analysis.html#hydrogen-bonding) , and (3) provides the option to read from LAMMPS trajectory (.lmppstrj) files.
Of the available HBond analysis functions in MDTraj, the Baker Hubbard (http://mdtraj.org/1.6.2/api/generated/mdtraj.baker_hubbard.html#mdtraj.baker_hubbard) function most closely matched the Hbond criteria employed by the group. We helped the group create a fork of the MDTraj code and modify the parts of the library required to reproduce the results of their own code. However – and to our surprise – we found the resulting code to perform much slower than the original Python code (for a 6.3 MB trajectory file, the initial code finished in about 18 seconds, while the code based on MDTraj took about 17 minutes). This large discrepancy is attributed to the fact that MDTraj checks each frame for bond properties whether they are wanted or not, while the original code looks for specific bond properties for the particular analysis the group is doing.
3. Using Hadoop/Spark
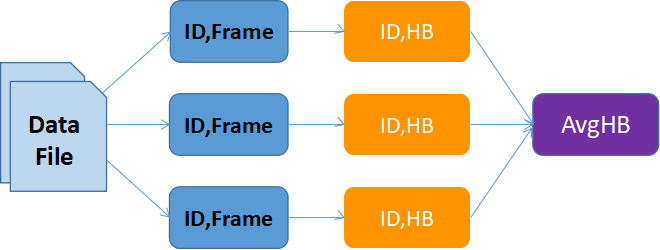
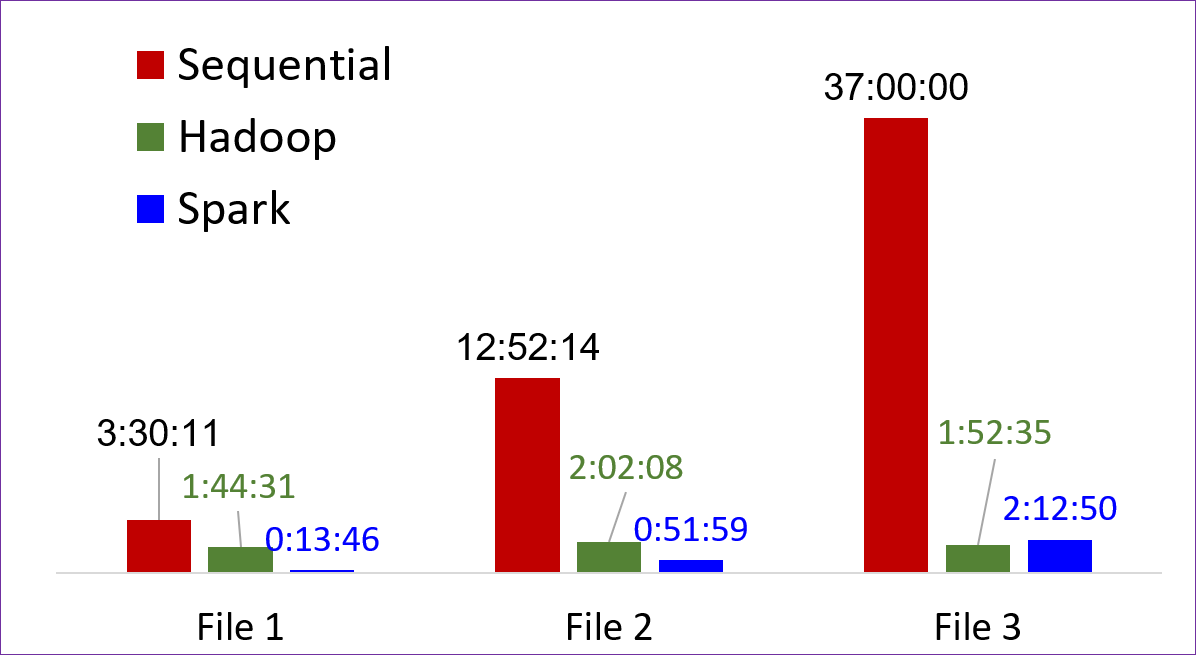
Since the trajectory files can be split into several frames that can be independently analyzed to count the HB per frame (Map) followed by theand the average HBond count can be found useing of an operation across the results from all files (Reduce) to determine the AvgHB), this is a very good fit for technologies such as Hadoop and Spark. A schematic of this workflow is shown in Figure 2. At Clemson University, researchers have access to a 3.64 PB Hadoop Cluster with 40 worker nodes, that can be exploited for performing these types of calculations. Starting with providing training on Hadoop/Spark, we worked with the group on developing Hadoop and Spark workflowsworfklows for parallelizing the calculation across several workers. Both approaches have their relative advantages and drawbacks. The Hadoop approach was found to be more tedious while slightly better performing for larger files, while the Spark workflow was more natural while still performing quite well. The resulting codes are muchseveral (up to 20) times faster than the original (fixed) sequential code, as shown in Figure 3.
The Result
After these steps, the resulting product is a code that performs much better than the initial code the group approached us with. Further, members of the group are trained in using Hadoop and Spark to post-process results from their simulations and are able to adapt the approach to future requirements. We also gained experience using the MDTraj library which, while not appropriate for this analysis, could be very useful for other use cases.
Supporting Media
Figure 1: Figure showing atoms at a particular frame and example of hydrogen bond (HB) that needs to be identified in the analysis
Figure 2: Data flow showing the map and reduce steps to compute the average HB
Figure 32: Time in hours to post-process three3 different trajectory files for the three methods described
Collaborators
- Rachel Getman, Professor, Chemical Engineering, Clemson University
- Yu Li, Ph.D. Candidate, Computer Science, Clemson University
- Xiaohong Zhang, Ph.D. Candidate, Chemical Engineering, Clemson University
- Linh Bao Ngo, Director of Data Science, CITI/ACI-REF, Clemson University
- Ashwin Srinath, Research Facilitator, CITI/ACI-REF, Clemson University
Funding Sources
- This research was funded by the National Science Foundation under grant numbers CBET-1554385. We also acknowledge financial support from Clemson University start-up funds.