Martin Cuma, University of Utah, August 30, 2018
Research computing support sometimes requires different twists and changes in direction. This was the case with enabling University of Utah Physics Professor Saveez Saffarian and his group to process iPALM (Interferometric photoactivation and localization microscopy) data on the University of Utah’s Center for High Performance Computing (CHPC) resources. Previous to their use of iPALM, the Saffarian lab data processing and imaging was carried out at a smaller scale on individual PC analysis machines in their lab. This was not practical for the iPALM experimental technique due to both the amount of data and the processing time needed, leading the Saffarian group to look to CHPC for assistance. Working with CHPC was critical to allow a full transition of all of the Saffarian group’s data storage and processing onto CHPC resources.
The Objective
The Saffarian group develops microscopy techniques that focus on live high-resolution imaging of essential molecular mechanisms critical for the function of retroviruses, such as HIV. As a part of this effort the group has been using the iPALM technique, which is capable of 3D imaging at a resolution of 10-20 nm. Specifically, iPALM has a greatly improved (about 2.5 times better) azimuthal resolution over the original PALM techniques that have been used by the group. At this resolution, protein molecules which constitute the virus can be observed in vivo, providing new insights into viral evolution and life cycle. However, with the increased resolution come larger data sets (10s of GB per image), which need to be processed by a custom software, developed at Howard Hughes Medical Institute (HHMI), to identify the objects of interest. As this analysis is time consuming, HHMI developed a cluster processing offload (i.e., a process to distribute the analysis workload to a high performance computing cluster instead of running it sequentially on one computer) via a Graphical User Interface (GUI) frontend.
Both the increased amount of data generated with the use of iPALM as well as the increased processing time associated with the new analysis software made the group realize that they needed to look beyond using their local resources to both store and process the data, and this prompted Professor Saffarian to contact CHPC to discuss the use of CHPC resources. After an initial meeting with Dr. Anita Orendt, the Professor met with CHPC Scientific Consultant and ACI-REF Dr. Martin Cuma, who, among other things, maintains Interactive Data Language (IDL), which is the language used for the iPALM data processing software. Getting the iPALM processing software to work was not a simple task, as described further below.
The Solution
As mentioned above, the iPALM analysis program, called PeakSelector, is written in IDL, short for Interactive Data Language, a programming language used for data analysis. Martin thought that he would install the program, interface it with the cluster scheduler and be done. However, using the IDL runtime that was already installed on our Linux clusters resulted in a segmentation fault, no matter what IDL version was used. Doing the same steps on a Windows system worked fine. As CHPC had been exploring containers at the same time, Martin tried to create a Singularity Windows container using the WINE emulator on Linux. This is quite atypical but it worked, although it ended up not being very stable and user friendly. Therefore, this lead Martin back to exploring the Linux error. As this error was not known to the HHMI developer, who was using the program in a Linux environment, Martin reached out to the IDL support which promptly suspected a known bug in a function that generates some of the GUI widgets and suggested that the GUI be rewritten. However, as the GUI is quite complex and the HHMI developer was focusing on other projects, rewriting the GUI would have resulted in a significant delay in the iPALM deployment.
The mystery remained why Peakselector works at HMMI; multiple calls with the developer did not provide any additional information as to the source of the differences. A few months later, Professor Saffarian and his graduate student, Ipsita Saha, were planning a trip to HHMI in Maryland to work with the iPALM developers on learning the technique, and invited Martin to join them for a few days. This turned out to be very productive on several fronts, as described below.
First, we saw with our own eyes that the program worked at HHMI, and by.analyzing the software and hardware being used it was found that the Linux crash was video related. After trying many different systems it was determined that the program works on a handful of Nvidia video cards. Once back home, we tried a number of existing machines with the Nvidia video cards that we knew worked at HHMI, with mixed success. The crashing mystery continued. To extend the pool of machines that we had available for troubleshooting, we contacted the University Bookstore and spent a couple of hours with their extremely accommodating staff to try a wide range of computers, and finally found the magic solution – an Nvidia video card paired with ultra-high-definition (higher than 1080p) resolution display.
In addition to determining the source of the crash, the remainder of the stay at HHMI was spent getting to know the HHMI cluster setup, and implementing the HHMI Peakselector cluster offload at CHPC. Peakselector divides that analysis into hundreds of independent serial calculations each of which is submitted to a cluster. HHMI allows node sharing, but CHPC does not, so, apart from modifying the submission system from LSF scheduler to SLURM, we also had to come up with a strategy to aggregate the individual calculations into groups to fill up all the cores on CHPC cluster nodes. Compared to the GUI crash troubleshooting, this was the easy part.
Once we had a stable platform on which to run the Peakselector, as well as the strategy to run the calculations on CHPC resources, the rest of the onboarding was quite standard. Martin helped Professor Saffarian and Ipsita set up the FastX remote desktop program on their local computers. He also installed and created the module file for Peakselector on CHPC’s Linux systems and taught the Saffarian group how to run Peakselector and how to monitor batch jobs. Without the transition to CHPC resources, the processing of the iPALM data on the existing Saffarian group desktops would have been slow if not impossible. In addition, without the technical assistance of ACI-REF Martin Cuma, the group would not have been able to make this transition.
With the move to using CHPC resources for the analysis also came the need to purchase storage (CHPC’s group space offering) to address the larger amounts of data that the group was now generating. In addition, the transfer of the data from the microscope to this storage needed to be explored. Therefore, along with engaging Martin, we also worked with the the CHPC networking team to explore the network connection options in the new lab space that will be hosting the iPALM microscope, and worked with campus networking to arrange for a 10Gb/s connection (originally set for a 1GB/s connection) from the instrument to the campus backbone. This connection allowed for the timely transfer of the images from the microscope to the newly purchased CHPC group space for storage and processing.
The Result
The newly installed iPALM software was used to finish the analysis of preliminary iPALM data collected in summer of 2017 and resulted in manuscript on the development of iPALM microscopy, “Correlative iPALM and SEM resolves virus cavity and Gag lattice defects in HIV virions,” which is under review. The figure below shows the clear resolution advantage of iPALM versus the microscopy technique previous used to study these type of systems.
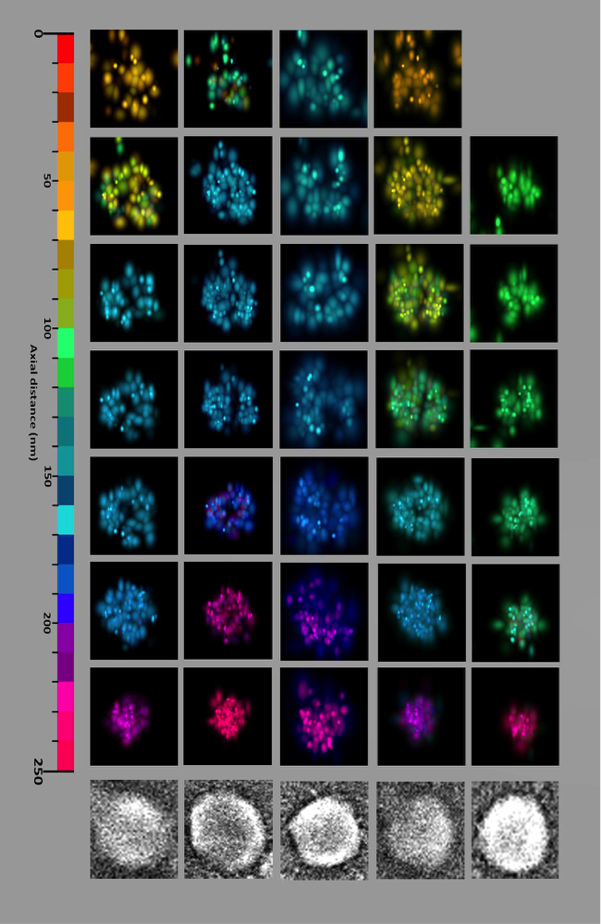
The Saffarian Lab was awarded an NIH RO1 award in summer of 2018, and their initial work with iPALM microscopy on purified HIV virions and cellular pathways was an essential component in obtaining this award.
From CHPC’s standpoint the main take home message of this exercise was that things are not always clear cut and that troubleshooting and finding alternatives is a part of our support job. In addition, the project was a learning experience for Martin as he got the chance to explore different alternatives for IDL runtime deployment and got to see how research is supported and done at one of nation’s leading private research institutes (the HHMI).
Collaborators and Resources
- Saveez Saffarian, Ph.D., Associate Professor, Department of Physics, University of Utah
- Ipsita Saha, Graduate Researcher, Department of Physics, University of Utah
- Martin Cuma, Ph.D., CHPC Scientific Consultant and ACIREF
- Gleb Shtengel, Ph.D., Senior Scientist, Howard Hughes Medical Institute
Funding Sources
This study was supported by NIH R01GM125444 to Saffarian. Martin Cuma was supported in part by a grant from the National Science Foundation, Award #1341935, Advanced Cyberinfrastructure – Research and Educational Facilitation: Campus-Based Computational Research Support.